| Merging structures
One useful feature of MOLDRAW is the possibility of merge different
structures in one single structure. To achieve that go to
(File--->Merge mol file...) menu and then select the
structure you want to merge with the one actually on the screen. The new
structure is set in 3D space to a distance of 5 Angstrom to the righmost
position of the actual structure. This option is useful when a small molecule
has to be placed on a surface of an inorganic material and one would like to
export the final result to CRYSTAL for further quantum mechanical
calculation.
Below it is shown the case of a calcite (CaCO3) surface in which the alanine
molecule has to be added as an adsorbate. Because the Merge option keeps, as a reference, the
unit cell of last loaded structure, the sequence is the following:
|
|
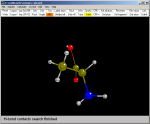
1) Read the file
r-ala.mol
in the usual way. Because this is a molecule the unit cell is simply 1
1 1 90 90 90. Click here for the alanine mol file. |
|
|
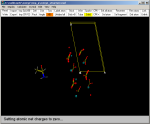
2) Then Merge the
calcite.mol file and select the
(Crystal--->View cell border) option. The cell is that
of the calcite surface. Click here for the calcite mol file. |
|
|
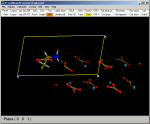
3) Select the Sel fragment
option of the toolbar. Choose the
fragment number 1 (alanine) and move it using the
usual mouse gestures until properly positioned.
See the Manipulation fragment
option to move/rotate molecular fragments resulting from the merge
process. |
|
|
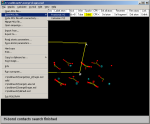
4) The resulting structure is then saved (Save--->Mol
File--->Fractional) in a new file which containg the merged
structure with the proper orientation and unit cell parameters. Choose to
save all atoms, even those outside the unit cell borders. Click here for the merged
file. |
|
|
5) The resulting merged structure is then read normally and by means of
the Pack option of the toolbar a large piece of crystal can
be generated. |
You are here: Home-Navigate-Topics-Merging
Previous Topic: Saving Next Topic: Perspective
|